It is mainly based on the biological affinity (or) biological specificity. This technique mainly requires previous knowledge of the molecule to be separated a specific ligand will only attach with a specific molecule.
The materials to be isolated are capable of binding reversibly to a specific ligand i.e., attached to an insoluble matrix.
Why Use Affinity Chromatography?
Affinity chromatography offers high selectivity, resolution, and capacity in most protein purification schemes. It is the only technique that has the advantage of utilizing a protein's biological structure or function for purification. As a result, purifications that would otherwise be time consuming and complicated, can often be easily achieved with affinity chromatography.
Supporting matrix:
1) Characteristics of Matrix:
- The matrix should be inert to other molecules to minimize non-specific adsorption.
- It should possess good flow properties.
- It should be chemically and mechanically stable at varying pH, ionic strength and denaturating conditions employed for binding and elution.
- It should contain large numbers of suitable chemical groups for ligand attachment.
- It should be highly porous a large surface area for attachment of the ligand and allows interaction of the desired macromolecule with the immobilized ligand.
2) Types:
The particles, which are uniform, spherical and rigid, are used. The most commonly used ones are
a) Agarose b) Polyacrylamide c) Controlled glass beads
a) Agarose:
The agarose beads have most desired features as mentioned above. But it has some advantage when use the denaturant solution for elution, which have a susceptibility to contraction.
b) Polyacrylamide:
The polyacrylamide bead lacks porosity. This undesirable trait is heightened even further when they are substituted by ligands.
c) Controlled porosity glass beads:
This bead provides mechanical rigidity and chemical inertness in addition to providing very good flow rates. High degree of nonspecific protein adsorption is the most serious drawback to these beads, which avoid to some extent by treatment with “Hexamethyldisilazane”.
3) Ligand selection:
The selection of ligand should have two most important requirements:
- Ligand interaction should be less with desired macromolecules.
- The ligand should possess functional groups that can be modified to form covalent linkage with the supporting matrix.
4) Ligand attachment:
Covalent coupling of the ligand to the supporting matrix involve the following steps:
- Activation of the matrix functional groups
- Covalent attachment of the ligand to the activated functional groups.
i) Activation of the matrix functional groups:
The most common method of activation of polysaccharide supports (agarose) involves treatment with “CNBr” at alkaline pH (pH=11.0). Usually 300 mg of powdered cyanogens bromide used per ml of packed gel gives the maximum substitution. The reaction is exothermic and maintains the temperature constant at 200C at all times. To maintain temperature the pH at 11, the mixture is continuously stirred and an electrode dipped into it at all times. The pH is maintained by the addition of 2M NaOH. The activated suspension is now washed with about 20 times the gel volume with a buffer (buffers –Tris, Ammonium acetate, Glycine) at a pH of 9.5 to 10. Usually just 10 to 15 minutes are required for the reaction to be completed. Sodium bicarbonate and borate buffers are the usual choice.
ii) Covalent attachment of the ligand to the activated functional groups:
Coupling of amino – containing ligand to CNBr activated support is normally carried out by suspending the support and the ligand in a basic buffer solution at pH (0.25 M NaHCO3, pH-9.0). The suspension stirred overnight in a cold room. During this time the ligand is covalently attached to the support medium.
After the reaction is over, the matrix should wash with 0.1 M solution of pH 9.0 glycine buffer, the solution destructs the any extra-activated groups.
The number of ligand group bound is usually expressed in terms of capacity per ml of packed matrix rather than in terms of its dry weight.
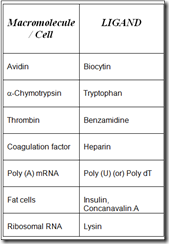
Some group specific ligands
5) The ARM:
To avoid the encounter steric repulsion between ligand and activated groups of matrix with macromolecule, which is used to introduce a spacer between the activated groups of the support and the ligand. This space is known as “ARM”. The ligand projects out the macromolecule to prevent repulsion.
E.g.:
1)Hexamethylene, 3,3’-diamino propylamine
2) 1,6-diamino hexane
3) 6-amino-hexanoic acid
4) 1,4-bis-(2,3-epoxypropoxy) butane
These spacer arms have two different functional groups; one to react with the functional groups of the matrix and the other is to react with ligand.
In organo synthetic procedures “Succinic anhydride” and a “Water soluble carbodiimide” are using to attaches the ligand.
Practical Procedure:
It is also carried out in the column. In this matrix and ligands are used. Prior to use, the gel (or) matrix must be converted to the swollen form, done by allowing a known weight of the gel to swell either in water (or) in a weak salt solution. The greater the porosity, the more will be the time required to reach equilibrium.
Agarose reacts with cyanogens-Bromide and forms activated complex. Then this activated complex reacts with epoxide and forms agarose cyanogens bromide-epoxide complex. Epoxide is a spacer arm attaches the matrix with ligands.
The agarose-CNBr-epoxide complex is filled in column. Then the specific ligand is added. Now the ligand react with the spacer arm and attaches to it. Then the sample is applied to the column. The specific ligand attaches with specific molecules. The remaining material will comedown the column, the attached molecule can be obtained by suitable buffer. The buffer supplement on the gel bed, it encourages adsorption of the desired molecule the buffer chosen must be supplemented with any cofactors (e.g.: Metal ions) required for “Ligand-Macromolecule interaction”. The buffer should also possess a high ionic strength so as to minimize non-specific polyelectrolyte adsorption onto charged groups in the ligand.
Applications:
- The technique has been used to purify a large variety of macromolecule such as enzymes, Immunoglobulins, membrane receptors, Nucleic acids and even polysaccharides.
- By using affinity chromatography, Whole cells have been purify include fat cells, T and B-lymphocytes, Spleen cells, Lymph node cells, Oocytes and chick embryo neural cells.
- Metal chelate affinity chromatography is the logical extension technique. Same molecular weight protein can be separated by this technique by using the metal ion containing matrix by chelation, because of their difference in their metal binding ability with proteins.
- By using the “Magnetic gel beads affinity chromatography”, immunoglobulin negative thymocytes and neuroblastoma cells have been purified by this method. The magnetic gel beads, usually polyacrylamide (or) agarose have a core made up of Fe3 O4 (Magnetite) and are chemically coupled to a protein ligand.
- Immobilized enzymes (Solid-state enzymes) are also isolated and purified by this method.
- mRNA can be isolated by this technique.
- Native proteins can be separated from denatured proteins by this technique.
- DNA & RNA can be separated from each other
- Papain and Urease can be separated by this technique.



No comments:
Post a Comment